HMW nuclear DNA extraction from difficult plant species for long read sequencing
In brief
Long read sequencing technologies require highly pure high molecular weight (HMW) intact DNA to utilise the optimal capacity of these technologies. Getting pure HMW DNA in non-model plant is a challenge due to complex cellular metabolites that chemically behave like DNA molecules. Isolation of nuclei helps to remove the majority of cellular impurities in DNA extraction processes. Recently, few protocols for HMW DNA extraction from plant species targeting to long read sequencing have been developed from different laboratories. Herein, the HMW nuclear DNA extraction method is optimised at the nuclei isolation step where the centrifugation speed is independent of genome size. Using this method, highly pure HMW nuclear DNA was extracted from two difficult plant species with a wide range of genome size – Macadamia jansenii (~800 mb) and Beauregard sweet potato (~2.4 Gb) and the quality was assessed by nanopore sequencing.
Highlights
-
Using this method, highly pure nuclear HMW DNA suitable for a long read sequencing can be extracted in 5-6 h from difficult plant samples.
- This described method does not require prior knowledge of genome size and optimisation of centrifugation speed during nuclei isolation therefore users can save optimisation time and materials while working with a new plant species.
- High throughput data (28 – 93 Gb in PromethION, 5-20 Gb in MinION) with long reads up to 295 kb has been generated in ONT using DNA prepared from this method.
Introduction
Long read sequencing technologies is now becoming popular in whole genome sequencing of a complex organism and other several genomics studies where short read sequencing technologies unable to provide genomic information(Jain et al., 2018, Li et al., 2017, Mantere et al., 2019, Paajanen et al., 2019, Sakamoto et al., 2020). Single molecules Real-Time (SMRT) sequencing from Pacific Bioscience (PacBio) and nanopore sequencing from Oxford Nanopore Technologies (ONT) are the most common long read sequencing technologies. Although these technologies offer advantages over short read technologies such as Illumina, DNA sample preparation is a challenge due to requirement of highly pure high molecular weight DNA (>50kb). Most of the existing commercial DNA extraction kits are column based that shear DNA into shorter fragments or suitable for limited species only. Therefore, conventional DNA extraction methods such as CTAB/phenol-chloroform (Doyle, 1991) are applied for HMW DNA extraction which can be modified according to the need of users. However, these methods require a lot optimisation or sometimes difficult to meet the requirements of DNA sample quality to long read sequencing. Recently, few protocols have been developed based on nuclear DNA extraction from different laboratories to eliminate most of the cellular impurities, but these methods require either handling of the high number of chemical reagents (Jones, 2019) or optimisation based on the genome size (Workman et al., 2018). Herein, Workman et al (2018) method is modified where the plant nuclei are first isolated from tissues preferably young and healthy followed by extracting HMW DNA using Nanobind Plant Nuclei Big DNA Kit (Circulomics, 2018). In the original method, centrifugation speed requires to be optimised based on the genome size during nuclei isolation which is a time consuming and sometimes difficult in plant species of unknown genome size. In this modified method, a high speed 3000×g is set up to pellet nuclei and other cellular debris that passed through miracloth filtration and a low speed 200×g to remove plant debris as a pellet. Thus, isolated nuclei can collect and wash at 3000×g as described in the original method. We successfully applied this method to extract HMW nuclear DNA in two plant species with a wide range of genome size, and compatibility of nuclear DNA samples in long read sequencing was tested in nanopore sequencing.
Materials & equipment
Materials
- Trizma Base (T6066-500G, Sigma)
- Potassium Chloride (P5405-500G, Sigma)
- EDTA (EDS-500G, Sigma)
- Spermidine trihydrochloride (S2501-1G, Sigma)
- Spermine tetrahydrichloride (S1141-1G, Sigma)
- Sodium hydroxide (1064980500, Merck)
- Sucrose (S0389-500G, Sigma)
- Triton X-100 (T8787-100ML, Sigma)
- β-mercaptoethanol (M6250-100mL, Sigma)
- PVP-360K (PVP360-100G, Sigma)
- Nanobind Plant Nuclei Big DNA Kit (SKU NB-900-801-01, Circulomics)
- Nuclease free water
- Ethanol (>98%)
- Liquid Nitrogen
- Homogenisation buffer (HB) stock 10x (see SOP for recipe)
- HB buffer 1x
- Triton X-100 (20% v/v, 100 ml) mix (see SOP for recipe)
- Nuclei Isolation Buffer (NIB) (100ml for 5gm tissue materials and prepare this buffer on the day of experiment) (See SOP for recipe)
Equipment and consumables
- Wide bore pipette tips (P1000 and P200)
- Falcon tubes (50 ml and 15 ml)
- Spatula
- Beaker (250 ml)
- Temperature controlled Centrifuge
- Mortar and pestle
- HulaMixer
- Magnetic rack (1.5 ml)
- Microscope (Florescence &/bright field)
- Bucket for transporting liquid nitrogen
- Magnetic rod and stirrer or equivalent
- Nanodrop
- Qubit
- Miracloth (US1475855-1R, Merck)
Procedure
Note : Please refer 'UQ_GIH_HMW_Plant_nDNA_extration.pdf' in the side for full details of the procedure, including 'critical points' and ' Occupational Health and Safety'.
A. Nuclei isolation
- Grind 5 gm of healthy leaves (fresh or fresh snap freeze) in liquid nitrogen into fine powder using mortar and pestle. Grinding takes 20-30 min to make fine powder and topping up of liquid nitrogen 4-5 times.
- Transfer the grinded leaves powder in a 500 ml beaker containing 50 ml (for 5 gm) ice-cold NIB and stir with magnetic stirrer for 15 min. Complete disruption of leaves powder clumps is essential for the better recovery of nuclei.
- Gravity filter (20 min) the leaves powder solution through 5-layers of Miracloth in a cold room and collect the filtrate in 50 ml falcon tube.
- Spin the filtrate at 3000×g for 20 min at 4°C and discard the supernatant. Resuspend the pellet in 1-2 ml ice-cold NIB buffer using wide bore 1ml pipette tips and adjust the final volume of 45 ml with ice-cold NIB buffer.
- Spin the resuspended solution at 200×g for 15 min at 4°C and transfer the supernatant in a fresh 50 ml falcon tube. Spin the transferred supernatant at 3000×g for 15 min at 4 °C and discard the supernatant.
- Resuspend the pellet in 1 ml of ice-cold NIB with the help of 1 ml wide bore pipette tips. Transfer the resuspended solution in a 15 ml falcon tube and make the final volume of 15 ml with ice-cold NIB. Mix the solution by inverting the tubes 5-6 times in cold room.
- Spin the solution in 3000×g for 10 min at 4 °C. Discard the supernatant and resuspend the pellet with 15 ml ice-cold NIB.
- Spin the solution at the same condition in step 7 and repeat this step 3-4 times or until the supernatant appears as a clear solution.
- Resuspend the pellet with 1 ml ice-cold 1×HB buffer. Keep 20 µl to observe isolated nuclei under microscope (20× and 40×). Spin the remaining solution at 5000×g for 5 min and discard the supernatant. Snap freeze the pellet in liquid nitrogen and store at -80°C.
B. DNA extraction using Nanobind plant nuclei big DNA kit (Circulomics)
- Resuspend the nuclei pellet directly in 60 µl of proteinase-K and vertex until it becomes a homogenous mixture. Vertexing in this step is very important for proper lysis of nuclei and getting pure DNA.
- Add 15 µl of RNase and vertex 10× and incubate at RT for 5 min to remove RNA.
- Add 160 µl of AP1 buffer and vertex 5× each for 2 sec. Vertexing of sample is necessary for the proper lysis and getting pure DNA.
- Incubate the sample in a thermomixer at 55 °C, 900×g for 2 h. Check every 30 min and mix the sample by inverting the tubes 5-6 times and a quick spin down for 2 sec and continue incubation.
- Spin the sample at 13,000×g for 5 min at RT and transfer the supernatant into a 1.5 ml Eppendorf tube containing Nanobind disk. Use wide bore pipette tips from this step. Adding Nanobind disk prior transferring the supernatant help to merge disc in the solution.
- Add equal volume of 100% Isopropanol and spin the tube in Hulamixer for 20 min. Set the parameters of Hulamixer as: 9 rpm rotation, tiling for 70° for 12 sec and vibration 2° for 1 sec.
- Place the tube in magnetic rack as recommended (Circulomics, 2018), Page-11.
- Discard the supernatant and avoid contacting Nanobind disk.
- Add 500 µl of Buffer PW1, inversion mix 4×, replace the tubes in the magnetic rack and discard the supernatant. Repeat this step once.
- Remove any residual liquid from cap if tube by spinning the tube on a mini centrifuge for 2 sec. Repeat this step. If the Nanobind disk is blocking the bottom of the tube, gently push it aside with the tip of the pipette. At this stage, DNA is tightly bound to the disk.
- Add 50-200 µl Buffer EB and spin the tube on a mini centrifuge for 2 sec. Incubate at RT for 10 min. Confirm the entire Nanobind disk is fully immersed in the Buffer EB during incubation.
- Collect DNA by transferring eluate to a new 1.5 ml Eppendorf tube. Use standard P200 pipette tip to remove residual liquid after much of the eluate has been removed with a side-bore pipette.
- Spin the tube containing the Nanobind disk on a mini centrifuge for 5 sec and transfer any additional liquid that comes off the disk to the previous eluate. Repeat it if necessary. A small amount of white or gel like material may remain on the Nanobind disk after transferring the eluate in a step 12. This is HMW DNA that has not fully solubilised and this spin step will allow DNA to slide off the Nanobind disk into the bottom of the tube, after which it can be pipetted out and combined with the previously transferred eluate.
- If the extracted HMW DNA is heterogenous and insolubilised, pipette mix 1-5 times with a standard P200 pipette tip and let the sample rest for overnight at 4 °C.
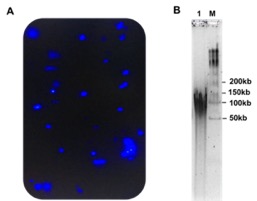
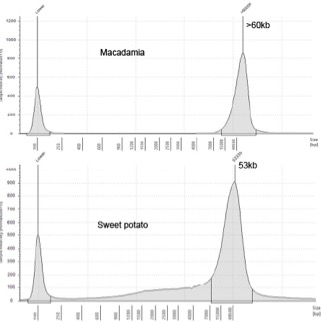
- Analyse the recovery and purity of the DNA by NanoDrop and Qubit. The ratio A260/A280 and A260/A230 should be 1.6-2.0 and >1.8 respectively. Analyse the DNA integrity in TapeStation and the distribution of DNA size in Pulse-Field-Gel-Electrophoresis.
Anticipated results
40-50 µg /5 gm wet healthy leaves (mixture of young and mature) with DIN value >8 and molecular weigh 50-150 kb. NanoDrop ratio A260/A280 1.8-2 and A260/A280 >1.8.
| Materials | gDNA yield | DNA average length (Kb) | DIN value | NanoDrop A260/A280 | NanoDrop A260/A230 |
|---|---|---|---|---|---|
|
M. jansenii leaves |
10 µg/gm |
50-150 (PFGE), >60 (TapeStation) |
9.4 | 1.92 | 2.2 |
| Beauregard sweet potato leaves |
3 µg/gm |
53 (TapeStation) | 8 | 1.81 | 1.95 |
| Sample | Macadamia jansenii | Macadamia jansenii | Beauregard sweet potato | Beauregard sweet potato |
|---|---|---|---|---|
| Sequencing device | PromethION | MinION* | PromethION | PromethION |
| DNA Shearing | No | No | No | NO |
| Size selection | SRE kit | No | SRE kit | No |
| ONT Chemistry | LSK109, R 9.4.1 | LSK109, R 9.4.1 | LSK109, R 9.4.1 | LSK109, R 9.4.1 |
| Nuclease flush | No | No | Yes (1) | No |
| Run duration | 64 h | 48 h | 64 h | 48 h |
| Sequence yield (Gb) | 28.3 | 4.97 | 93 | 19.6 |
| Mean read length (kb) | 14.3 | 3.7 | 5.9 | 4.2 |
| Max read length (kb) | 295 (QC>9) | 140 (QC>9) | 165 (QC>9) | 128 (QC12) |
| Read length N50 (kb) | 33.3 | 9.5 | 13.4 | 10 |
*DNA sample was from a different lot
Troubleshooting
- Hard to grind tissues: mixture of healthy young and mature or healthy young (better yield and easier to grind). if leaves are the starting tissue, remove the leaves vein before doing snap frozen or direct grinding.
- Difficult to homogenise tissue powder: Make sure tissue powder does not contain liquid nitrogen. A good idea is to keep mortar containing tissue powder on dry ice and let it dry. Use the higher volume of NIB buffer.
- Low amount of nuclei Isolation: Use young tissues and squeeze the miracloth once the gravity flow completed to get all the nuclei those stuck in miracloth. Use wide-bore pipette or brush while resuspending the pellet in order to prevent the nuclei from physical damage.
- Troubleshooting regarding DNA extraction: see the manufacturer’s troubleshooting guide (Circulomics, 2018), Page-21-23
References
CIRCULOMICS 2018. Nanobind Plant Nuclei Big DNA Kit Handbook v0.18.
DOYLE, J. 1991. DNA Protocols for Plants. In: HEWITT, G. M., JOHNSTON, A. W. B. & YOUNG, J. P. W. (eds.) Molecular Techniques in Taxonomy. NATO ASI Series (Series H: Cell Biology) ed. Springer, Berlin, Heidelberg.
JAIN, M., KOREN, S., MIGA, K. H., QUICK, J., RAND, A. C., SASANI, T. A., TYSON, J. R., BEGGS, A. D., DILTHEY, A. T., FIDDES, I. T., MALLA, S., MARRIOTT, H., NIETO, T., O'GRADY, J., OLSEN, H. E., PEDERSEN, B. S., RHIE, A., RICHARDSON, H., QUINLAN, A. R., SNUTCH, T. P., TEE, L., PATEN, B., PHILLIPPY, A. M., SIMPSON, J. T., LOMAN, N. J. & LOOSE, M. 2018. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol, 36, 338-345.
JONES, A., BOREVITZ, J. 2019. Nuclear DNA purification from recalcitrant plant species for long-read sequencing V.3 https://www.protocols.io.
LI, C., LIN, F., AN, D., WANG, W. & HUANG, R. 2017. Genome Sequencing and Assembly by Long Reads in Plants. Genes (Basel), 9.
MANTERE, T., KERSTEN, S. & HOISCHEN, A. 2019. Long-Read Sequencing Emerging in Medical Genetics. Front Genet, 10, 426.
PAAJANEN, P., KETTLEBOROUGH, G., LOPEZ-GIRONA, E., GIOLAI, M., HEAVENS, D., BAKER, D., LISTER, A., CUGLIANDOLO, F., WILDE, G., HEIN, I., MACAULAY, I., BRYAN, G. J. & CLARK, M. D. 2019. A critical comparison of technologies for a plant genome sequencing project. Gigascience, 8.
SAKAMOTO, Y., SEREEWATTANAWOOT, S. & SUZUKI, A. 2020. A new era of long-read sequencing for cancer genomics. J Hum Genet, 65, 3-10.
WORKMAN, R., TIMP, A., FEDAK, R., KILBURN, D., HAO, S. & LIU, K. 2018. High Molecular Weight DNA Extraction from Recalcitrant Plant Species for Third Generation Sequencing. Protocol Exchange.
Associated project
Publications
Related development
Standard operating procedure (SOP)
Please contact Stacey Andersen (operations manager) for the updated SOP.
GIH team
Collaborating researchers