Fourth edition 2021 - Director's note

Dear Colleagues,
Welcome to the fourth edition of the GIH newsletter for 2021. Firstly, in this issue we are pleased to introduce our new Deputy Director, Dr Allan McCrae.
We also announce our 2022 External Project call, the GIH team's nomination for the UQ Staff Excellence Awards and congratulate GIH's collaborators and committee members on recent NHMRC grant success. We highlight one of our collaborative projects, our latest publications and seminars. We shine a spotlight on one of project leaders, Dr Denuja Karunakaran as well as our collaborative computing facility QCIF Bioinformatics.
We hope you enjoy the read. This is the last edition for 2021. The next edition of the GIH newsletter will be distributed in February 2022.
Professor Grant Montgomery, Director
Deputy Academic Director appointment

The position of a GIH Deputy Director was advertised earlier this year and we are pleased to advise that Dr Allan McRae has accepted the position.
Allan has an ARC Future Fellowship and is a Group Leader at the Institute for Molecular Biosciences at The University of Queensland.
Alans speciality is in the field of Systems Genomics. He has experience in agriculture and human studies and also with a range of genomic technologies, including genotyping arrays, sequencing, transcriptomics, epigenetic and microbiomics.
We would like to welcome Allan to the Management Group and look forward to his ongoing contributions to GIH.
Call for 2022 GIH External Project applications
In 2018, GIH began engaging in external collaborative projects in partnership with research groups across UQ to develop genomic-based cutting-edge technology breakthroughs (methods and pipelines). Collaborating research groups work closely with GIH in the design and development of projects as well as actively contributing to projects, including co-investment in funding and personnel expertise. Our projects are prioritized on the basis of novelty and transformative impact in advancing genomic applications and/or those that significantly drive down the costs of these applications.
2022 External Collaborative Project call -now open!
Collaborating research groups with proposals for innovative genomic projects will be again sought for 2022. For successful applications, GIH staff and potential funding toward GIH consumables will support each project to develop cutting-edge technology breakthroughs (methods and pipelines). Proposals can be entirely wet-lab-based, entirely bioinformatics-based or a combination of the two, and will vary in requirements for GIH budget and/or GIH staff support.
To find out more, head over to our specific 2022 GIH External Project Call webpage here.
2022 GIH External Project Call
2021 UQ Awards for Excellence

The GIH team was proud to be nominated for the 2021 UQ Awards for Excellence!
On the 22nd October, we celebrated the outstanding work of staff working at all levels across the University at an official ceremony at the UQ Centre. The UQ Awards for Excellence celebrate UQ achievements over the past year, acknowledging the outstanding contributions, innovative work and exemplary leadership demonstrated by staff at UQ. We congratulate all the 400+ staff associated with nominations for the 2021 UQ Award for Excellence!
We've had a great year working with all our colleagues including everyone within our collaborative projects and would like to thank our nominators, supporters and researchers for contributing to our success!
Visit the Staff Awards webpage to find out about the fantastic efforts of all everyone nominated and the chosen awardees.
2021 NHMRC Grant Success!
We are thrilled to report some of the recent NHMRC funding successes received by some of our fantastic UQ collaborative researchers!
Dr Quan Nguyen Senior Research Fellow, Group Leader, Institute for Molecular Bioscience) has been awarded a $1.35 million Investigator Grant titled "SPatially ACcurate Evaluation (SPACE) of Cancer Biopsies". His current GIH 2021 project "Developing new spatial proteomics capability and extending the applications of spatial transcriptomics to clinical archival tissue samples" aims to provide UQ researchers with access to cutting-edge expertise in “spatial omics”, which could be applied across all tissue specimens.
Dr Denuja Karunakaran (Group Leader, Institute for Molecular Bioscience), has been awarded a $1.27 million Ideas Grant titled "Inflammatory Cell Death, Circadian Rhythm & Obesity: A Surprising Triad". Denuja is this issue's featured collaborator who is leading the 2020 GIH Project Combining novel scSLAM-seq technology with 10x Genomics Chromium to track microchanges in newly synthesised RNA. Scroll down to our Collaborator spotlight to read more about her research project with GIH.
Dr Rodrigo Suarez (Lecturer (Anatomy and Physiology), School of Biomedical Sciences, Faculty of Medicine) secured a $0.88 million Ideas Grant titled "Building the mind: interplay of transcriptional and electrical activities in the self-assembly of cortical circuits in vivo". Rodrigo is part of the exciting 2021 GIH project "Utilising RNA velocity quantification of single cell transcriptomics to study differential cellular ontogeny in a single stage sample across related species." Along with his collegues, Rodrigo aims to develop a new bioinformatic tool to advance the study of the roles of developmental timing on complex trait formation. Furthermore, they hope the scripts can also be applied to the study of disease and be used to perform comparative analysis of mRNA maturation dynamics across disease and control conditions.
Dr Christian Nefzger (Senior Research Fellow, Institute for Molecular Bioscience) was awarded $0.77 million for "Reinforcing the cell identity network to improve the function of aged cells". Christian is driving the 2021 GIH Project "Expanding the scope of multimodal single cell sequencing" where the aims include to develop a workflow for sample multiplexing via antibody based indexing of nuclei, and establishing a robust analysis pipeline for sample demultiplexing of the sequencing data.
We also congratulate NHMRC funding successes received by members of the GIH Steering Committee!
Dr Kirsty Short (Senior Lecturer, School of Chemistry and Molecular Biosciences, Faculty of Science) was awarded a $1.37 million NHMRC Investigator Grant for "Pandemic-proofing our future".
Prof Philip Hugenholtz (ARC Australian Laureate Fellow, School of Chemistry and Molecular Biosciences, Faculty of Science and Affiliate Professor, The University of Queensland Diamantina Institute, Faculty of Medicine) was awarded $0.73 million Ideas Grant funding for "Dual-function ribonucleases: unexpected agents of antibiotic resistance".
Development: CRISPR-Cas9/Cas12a protein expression, purification and validation
CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated) technologies enable scientists to accurately modify the genome of cells and organisms.
CRISPR-Cas9 and CRISPR-Cas12a enzymes are the most widely studied of the RNA-guided endonucleases used for genome editing.
To help reduce costs of our genome editing research projects, we have established an "in-house" protocol for purifiying and functionally testing a range of Cas9/Cas12a endonucleases which are attached to various affinity tags. Our "In-house" purified Cas9 and Cas12a show comparable quality with commercial available products.
The full protocol for CRISPR-Cas9/Cas12a protein expression, purification and validation is available on our GIH website.
Single-cell transcriptome and chromatin profiling in plant cells
Transcriptional and chromatin profiling of single plant cells/nuclei is in its infancy. While great technical and analytical strides have been made in the field of single cell profiling, little of this capability has been translated to plant single cell profiling.
As a 2020 GIH collaborative project, "Single-cell transcriptome and chromatin profiling in plant cells", our main goal is to enable single cell plant functional genomics and systems biology across a variety of plant species (both model organisms like Arabidopsis as well as crops, e.g. wheat), research questions, and UQ units.
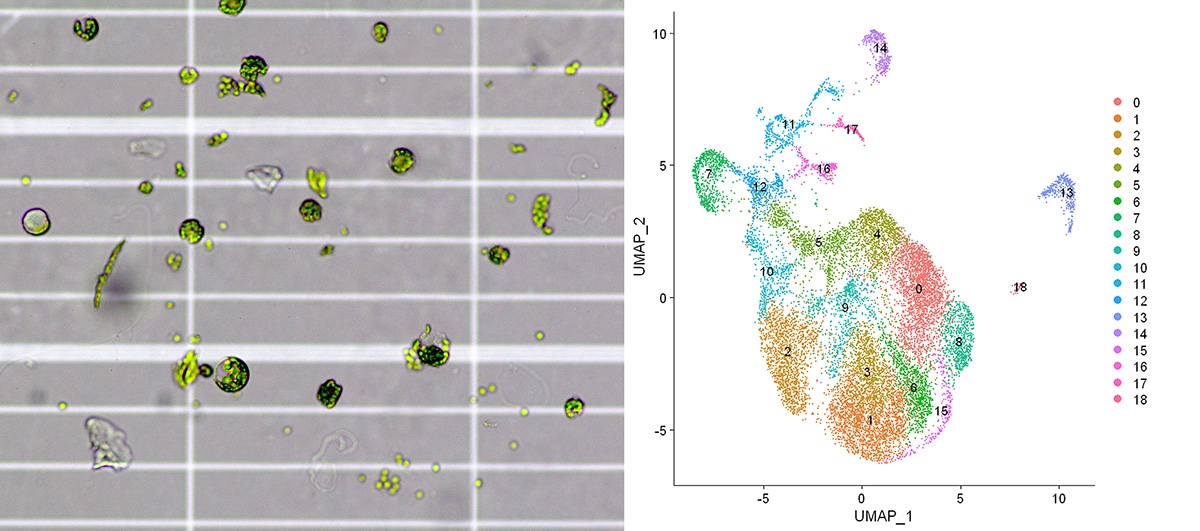
Transcriptional and chromatin profiling of single plant cells/nuclei is in its infancy. While great technical and analytical strides have been made in the field of single cell profiling, little of this capability has been translated to plant single cell profiling.
As a 2020 GIH collaborative project, "Single-cell transcriptome and chromatin profiling in plant cells", our main goal is to enable single cell plant functional genomics and systems biology across a variety of plant species (both model organisms like Arabidopsis as well as crops, e.g. wheat), research questions, and UQ units.
The aim of this project is to implement and optimize single cell RNA-seq and ATAC-seq (scRNA-seq and scATAC-seq) profiling methods in plant cells and associated bioinformatics data analysis pipelines to enable future discoveries in plant biology and agriculture across the large UQ plant biology research community. Led by group leader Dr Milos Tanurdzic (School of Biological Sciences, ARC Centre for Plant Research Success in Nature and Agriculture), with Dr Elizabeth Dun, Dr Franziska Fichtner and Dr Jazmine Humphreys and members of the GIH Team Dr Sohye Yoon, Stacey Andersen, Dr Subash Rai and Jun Xu.
We are nearing completion of this project so if you're interested in applying the developments to your research, have a look at the projects webpage and get in touch.
Read more about single-cell profiling in plant cells
GIH collaborator spotlight - Dr Denuja Karunakaran
Each newsletter we shine the spotlight on a lead researcher within one of our GIH collaborative projects.
We caught up with Dr Denuja Karunakaran, Team Leader at the Institute for Molecular Bioscience. She is a macrophage biologist with research interests in cardiometabolic diseases and therapeutics.
Denuja won the people's choice award for the 2020 Queensland Women in STEM Awards (see video above). Her research is leading a world study that uncovered an inflammatory gene responsible for obesity which has featured on local TV news networks.
With GIH, Denuja is leading the collaborative project "Combining novel scSLAM-seq technology with 10x Genomics Chromium to track microchanges in newly synthesised RNA". The aim of our project is to develop a novel pipeline for scSLAM-seq analysis combined with 10x Genomics Chromium to metabolically track transcriptional synthesis of new RNA during efferocytosis. This versatile technology can be applied to any model organism.
Can you tell us a bit more about your key research interests?
My research career has evolved around understanding and identifying novel therapeutic targets to treat cardiometabolic diseases such as atherosclerosis, obesity and diabetes. My research to date has defined novel pathways in platelet receptor regulation in thrombosis (Blood); macrophage inflammation, metabolism and cell death in atherosclerosis (Circulation Research, ATVB, Science Advances Circulation); and more recently, genetic regulation of adipose tissue inflammation during obesity (Nature Metabolism). My current research program aims to better understand how gene regulation of adipose tissue inflammation contributes to obesity, and in the longer-term how this in turn contributes to atherosclerosis.
Can you tell us a bit about your project with GIH and how this relates to your research?
Efferocytosis is a process by which dying cells are cleared, and our recent studies in Science Advances (2016) show that apoptotic cells are effectively efferocytosed unlike necroptotic cells. Although studies have demonstrated that transcriptional regulation of some specific genes occur during effective efferocytosis (i.e. clearance of non-inflammatory apoptotic cells), the precise single-cell transcriptional regulation during ‘defective’ efferocytosis (i.e. clearance of pro-inflammatory necrotic or necroptotic cells) remains to be elucidated and the focus of this project. Importantly, this project is developing novel SLAM-Seq technology to identify newly transcribed genes – first in Australia to the best of our knowledge
What is the goal of this project and how could it benefit other researchers?
Many studies rely on RNA-Seq data or similar to detect the differences between treated and non-treated cells, which includes basal and constitutive transcription. This new technology (scSLAM-Seq) enables us to detect very low levels of newly transcribed RNA in response to a specific stimulation or condition, providing novel insights into genetic response and regulation during specific homeostasis or disease processes.
Find out more about the scSLAM-seq project
Day in the life: QCIF Bioinformatics
QCIF Bioinformatics (QFAB) is a not-for-profit membership-based organisation that supports all the Queensland Universities. QFAB has a long running and constructive relationship with the University of Queensland, supporting its researchers through a variety of engagements including grant writing support, strategic advice, time-based projects, and embedded FTE positions. With GIH, QFAB supports UQ researchers through direct researcher interaction, a jointly employed bioinformatician with GIH (Valentine Murigneux) and by collaboration in research projects.
We caught up with the Head of Computational Biology at QFAB, Dr Gareth Price to find out more about a typical day at QFAB:

A typical day at QCIF Bioinformatics:
The Computational Biology team at QFAB is spread throughout Queensland, embedded at several of our member Universities (providing local support to that University).We are an agile organisation that is ready at a moments notice to support a researchers data science related question. For a researcher not being able to advance a project is time and money cost, so QFAB offers limited free advice to researchers to allow them to get past a minor roadblock in their analyses.
Importantly QFAB contributes as an equal partner to a number of national eResearch activities, under the Australian BioCommons, meaning my day is also interacting with the fantastic national skilled practitioners Australia has in the fields of eResearch, Supercomputing, Digital Infrastructure, Skill Development and importantly through to the scientists all these domains enable.
What do you all like best about your job:
My job at QFAB provides me the opportunity to interact with a vast array of researchers, both in their field-of-interest and the experience (students through to group leaders). I get, by proxy, to engage with these researchers through advice, grant support and project work. QFAB’s staff are scientists at heart, we enjoy enabling research and to help achieve the best outcome for the hard work that has gone into data generation. Getting the opportunity to plan for and enable our researchers to analyse the latest in -omics technologies is of great personal satisfaction to me.
Advice to others:
Get in touch and get in touch early! QFAB will always make the best of the data a researcher generates, however one of the best ways to maximise the opportunities in the data is to engage with us early. We can advise on grant support, experiment design, power calculations and end point analytical options. Our role is to get you to results quicker, so contact us for support here.
Make use of the UQ RDM and AARNet’s Cloudstor resources for long term data storage and secure data transfer options, respectively. Don’t keep your critical research data on a hard drive or USB stick.
Bioinformatics analyses is no different for wet-lab analyses, both benefit for good record keeping. Use a platform, like Galaxy Australia, to record all your analyses and make thesis and paper writing easier with one-click workflow and citation management.
To find out more about QCIF Bioinformatics (QFAB), click on the link below.
Latest publications
GIH is proud to support the research within the 2019 GIH project "Dual gRNA screen" led by our GIH collaborator Dr Nathan Palpant (Institute for Molecular Biosciences, University of Queensland).
We have generated a workflow for high efficient CRISPR knockout using optimised multiple guide RNAs design as part of this project. In association with the Queensland Facility for Advanced Genome Editing (QFAGE) and the Transgenic Animal Service of Queensland (TASQ), this workflow contributed to two recent excellent publications by UQ researchers.

Published in Circulation, by Dr Meredith Redd and colleagues as "Therapeutic Inhibition of Acid Sensing Ion Channel 1a Recovers Heart Function After Ischemia-Reperfusion Injury", their data provides compelling evidence for a novel pharmacologic strategy involving ASIC1a blockade as a cardioprotective therapy to improve the viability of hearts subjected to Ischemia–reperfusion injury .
Dr Di Xia used this workflow for the CRISPR transfection, troubleshooting and analysis required to generate the ASIC1a knock-out iPSCs utilised in the publication.
Published in the Journal of Cell Biology, Dr Harriet P. Lo and colleages as 'Cavin4 interacts with Bin1 to promote T-tubule formation and stability in developing skeletal muscle', they identified a role for the muscle-specific component, Cavin4, in skeletal muscle T-tubule development by analyzing two vertebrate systems, mouse and zebrafish.
In this research, Dr Di Xia utilised the CRISPR workflow for both the genome editing required to create both the Cavin4 Knockout mouse model and Cavin4 Knockout C2C12 cells.
Head over to our GIH website to find out more about the GIH Collaborative project "Dual gRNA" , led by A/Prof Nathan Palpant.
Check out all our collaborative publications and pre-prints
GIH developments and protocols
To find out about all our GIH developments from within our collaborative projects, click the links below.
Single-cell and short read sequencing | Imaging and spatial transcriptomics | Proteomics | Long-read sequencing | Bioinformatics and computational tools
Latest Bioinformatic Software: Methylartist
Congratulations to GIH collaborators Dr Seth W. Cheetham, Dr Michaela Kindlova and Dr Adam D. Ewing from the Mater Research Institute, University of Queensland, on their latest pre-print "Methylartist: Tools for Visualising Modified Bases from Nanopore Sequence Data" and the associated software release of the bioinformatic workflow Methylartist.
Methylartist is a consolidated suite of tools for processing, visualising, and analysing nanopore methylation data derived from modified basecalling methods. All detectable methylation types (e.g. 5mCpG, 5hmC, 6mA) are supported, enabling integrated study of base pairs when modified naturally or as part of an experimental protocol.
This amazing new software has generated quite a lot of interest on social media amongst the bioinformatics community is available to download via Adam's GitHub page.
For more information on Seth and Adam's 2020 GIH collaborative project with Professor Geoff Faulkner, see "Simultaneous identification of RNA-chromatin interactions and transcriptomes in single cells" on our website.
GIH collaborative research presentations
We've seen some great presentations from both the GIH team and our collaborators over the past few months. Here's a snapshot of some of them:

Prof Sandie Degnan and Prof Bernard Degnan, Faculty of Science, School of Biological Sciences, Centre for Marine Science, The University of Queensland
2021 GIH Project "Developing a scalable genome browser and repository for complex multi-omic datasets"
Genome annotation is crucial to defining the function of genomic sequences. Apollo is a popular tool for facilitating real-time collaborative curation and genome annotation editing. The Australian BioCommons and partners at QCIF and Pawsey are now offering the Apollo Service free to use for Australian-based research groups and research consortia.
As part of the launch, early adopters Prof Sandie Degnan and Prof Bernard Degnan, who have utilised the Apollo platform to provide the framework for their muti-omic browser in the GIH collaboratative project described what is possible with the tool. Their talk was recorded and is available to watch here.
Australian BioCommons is also holding a workshop making use of a training instance of the new Australian Apollo Service on the 17th November. More details are available here.
Find out more about this project

Professor Andrew Mallett, Professor of Medicine (College of Medicine, James Cook University) and Director of Clinical Research & Nephrologist (Townsville University Hospital & THHS), Clinical Fellow (Institute for Molecular Bioscience, The University of Queensland)
2019 GIH Project "Spatial genomics technologies to study cancer and genetic diseases in tissue contexts"
We were privileged to hear from Andrew Mallett, an Adjunct Senior Fellow and Nephrologist, about his research group's application of spatial transcriptomics in healthy and diseased kidney tissues. This spatial transcriptomics kidney project is part of an exciting and successful collaborative partnership between GIH, QIMR, TRI, Pathology Queensland and the Royal Brisbane and Women’s, Princess Alexandra, and Westmead Hospitals.
Find out more about this project

Dr Peter Crisp, School of Agriculture and Food Sciences, Faculty of Science, The University of Queensland
2021 GIH project "Enabling population scale epigenomics for crop improvement"
As part of the QAAFI Science seminar series, GIH collaborator, Dr Peter Crisp, a DECRA Research Fellow and Lecturer who leads a research group in the School of Agriculture and Food Science, described their recently developed approach that uses DNA methylation profiling of a single tissue (eg a leaf) to distill a genome down to the relatively small fraction of regions that are functionally valuable for trait variation throughout development. Peter's talk was recorded and available to watch here.
Find out more about this project

Dr Brooke Purdue, Genome Innovation Hub, The University of Queensland
As a speaker at the Oz Single cell conference, Brisbane node, Brooke presented an overview of the range of single-cell based projects that the GIH has undertaken over the last three years.
GIH developments presented included the bioinformatic tool scSplit for de-multiplexing scRNA-seq data and the optimisation of the RNAscope Hiplex assay for spatial transcriptomics. For more information on all our GIH developments, see our website here.
There was over 400 registrants at Brisbane Oz Single Cell conference. This year the theme was 'Cells in Space & Time' and brought together a collection of renowned speakers that covered themes including spatial transcriptomics, clinical studies, new tech and innovations, and data analysis challenges.
The Oz Single cells conference now moves onto Sydney on the 19th November, with the exciting theme 'Multiomics madness'! Click here to register.
Seminars, Workshops and Conferences
WORKSHOP: Refining genome annotations with Apollo, 17th November
This workshop will make use of a training instance of the new Australian Apollo Service. This service enables Australian-based research groups and consortia to access Apollo and host genome assembly and supporting evidence files for free. This service has been made possible by The Australian BioCommons and partners at QCIF and Pawsey. For details, see the Biocommons webpage https://www.biocommons.org.au/events/apollo-2021
UQ Hacky Hour, every Tuesday, 3–4pm.
UQs RCC and QCIF Bioinformatics teams are on hand to answer your computational questions every Tuesday afternoon via zoom. In addition, the GIH team are on hand to answer any genomic related questions on the first Tuesday of the month (Bioinformatics) and last Tuesday of the month (Experimental and sample prep). For Zoom meeting details please email: hackyhourUQ@gmail.com
QCIF training courses and workshops
QCIF (the Queensland Cyber Infrastructure Foundation) runs a number of training workshops throughout the year available to staff and students from six Queensland Universities including the University of Queensland. These include categories such as Bioinformatics Analysis, Data Processing and Statistics, Data sharing and reproducibility, Machine Learning, Programming / Software Carpentry, Surveys and Data Management, Visualisation and research communication. See the website for further information: https://www.qcif.edu.au/training/training-courses/
UQ Research Computing Centre Training and workshops
The UQ Research Computing Centre (RCC) runs a number of regular training sessions, specialty training sessions and on-demand training sessions with a focus on the computing resources available at UQ. See their training webpage for upcoming sessions and contact details UQ https://rcc.uq.edu.au/training#upcoming
UPCOMING SYMPOSIA AND CONFERENCES
OZ Single-cell 2021: Symposia series - Sydney, 19th November.
The single cell consortium comprises of leading researchers whose focus is on technologies that include single cell gene expression, genome sequencing, cell lineage tracing and the computational biology needed to tackle big data. Back by popular demand, and with a hybrid in-person (lockdowns permitting) and online attendance, the Oz Single Cell symposia series runs across the nation, across 2021. https://www.singlecells.org.au/
2021 Queensland Research Bazaar, 24-26 November
Hosted in 2021 by The University of Queensland, the 2021 Queensland Research Bazaar will be a three-day intensive conference where researchers will come together to up-skill in 'next generation digital research tools and skills'. In the spirit of a marketplace or bazaar, ResBaz is a highly participatory event where researchers from many different disciplines can learn, share knowledge and skills, and have fun! There's also aone-day satellite event, on Friday 26 November, at Griffith's Coast Gold Campus. https://resbaz.github.io/resbaz2021qld/
The Combined ABACBS and Phylomania 2021 hybrid conference, 22 - 26 November
The conference will be held in a hybrid setting this year, in person in the ‘hub’ site of each capital city and the talks will be video Zoomed LIVE across all states. Brisbane Hub coming soon, so keep an eye on their website for details. Their aim is to share cutting-edge bioinformatics and computational biology research, with an emphasis on how computational research is done. https://www.abacbs.org/conference2021
We'd love to know if you have found any of our GIH developments helpful for your own research, so drop us a line.
In addition to our quarterly newsletter, GIH news is also posted on our website here and on our Twitter feed here