Targeted capture and detection of 16s rRNA gene applying PCR free long read sequencing
Aim
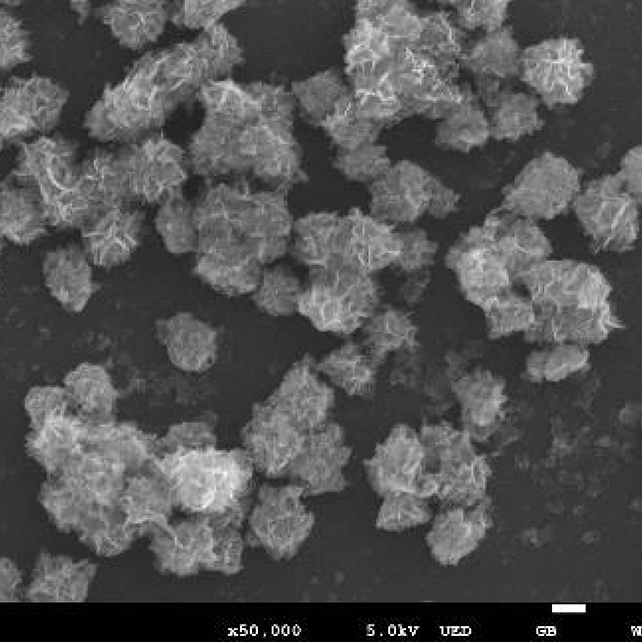
Use superparamagnetic nanoflowers to sequester bacterial 16s rRNA gene and apply long-read sequencing to identify and characterise low abundant bacteria.
Brief project outline
Our project utilises superparamagnetic nanoflowers to sequester the 16s rRNA gene from bacterial genome using sequence specific probes.
The probes will be conjugated on the nanoflowers using photocleavable linkers. This approach will enable us to specifically cleave the captured 16s rRNA gene for library preparation and subsequent nanopore sequencing. Sequencing the captured and cleaved fraction would enable us to directly sequence the gene without any need of amplification.
Genomics-based innovative aspect of proposal
Our project aims to utilize current capacities at GIH. GIH has Oxford Nanopore Sequencing capacities and expertise in method development. Leveraging the current capacities, we will introduce direct 16s rRNA gene sequencing. This new approach will also be a proof of principle to introduce specific gene of interest sequencing via nanopore platform without the need of gene amplification.
Broad applicability of the technique
The primary utility of this technique will be for diagnostics applications. The current proposed method will help researchers in the area of bacterial detection and microbial genomics. It provides an opportunity to elucidate base modifications in the 16s rRNA gene providing new information. If successful, this approach could be extended to detect antimicrobial resistance (AMR) genes in bacteria and, rare genes and target sequence of interest such as telomere.


