scSplit tool: genotype-free demultiplexing of pooled single-cell RNA-seq
 In brief
In brief
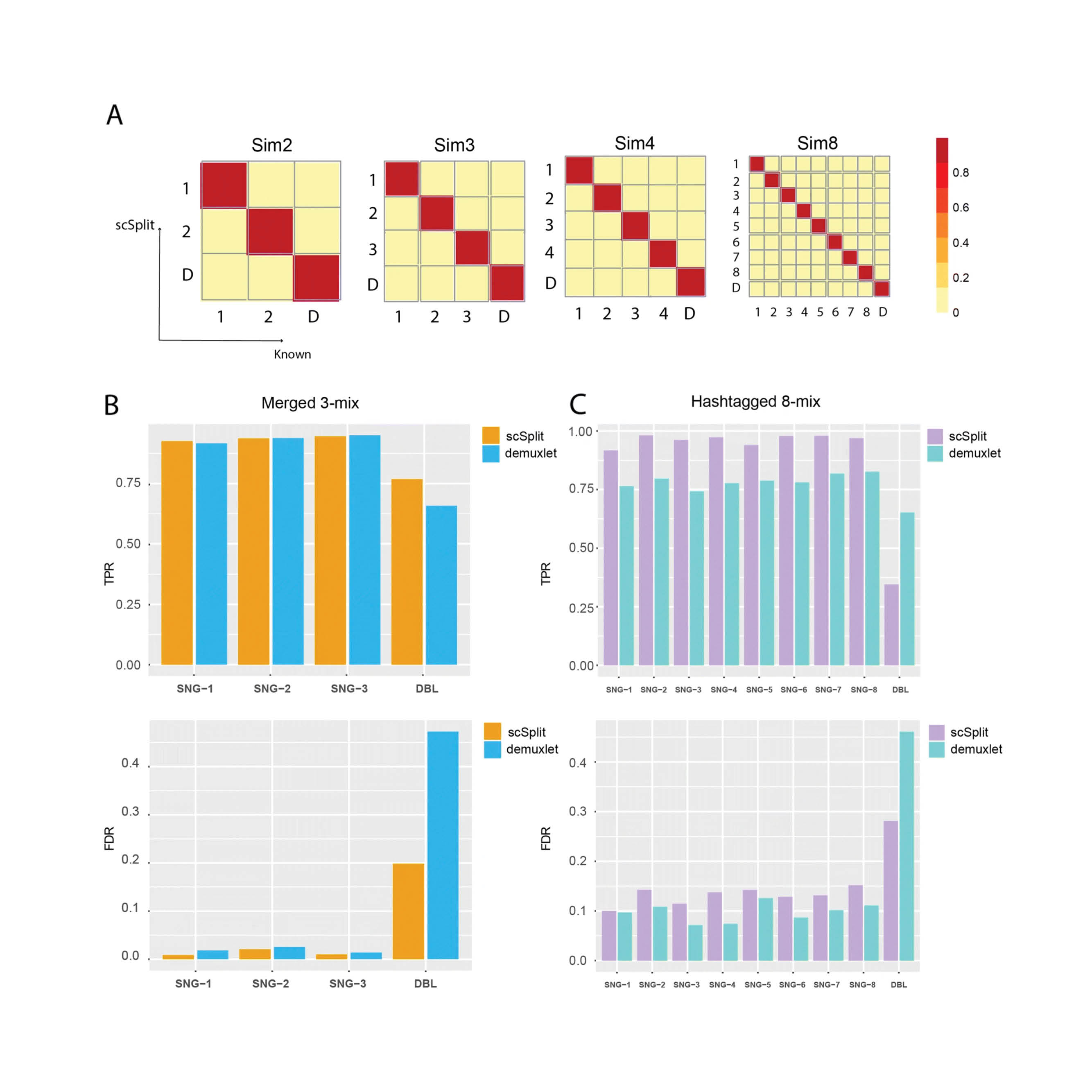
A variety of methods have been developed to demultiplex pooled samples in a single cell RNA sequencing (scRNA-seq) experiment which either require hashtag barcodes or sample genotypes prior to pooling. We introduce scSplit which utilizes genetic differences inferred from scRNA-seq data alone to demultiplex pooled samples. scSplit also enables mapping clusters to original samples. Using simulated, merged, and pooled multi-individual datasets, we show that scSplit prediction is highly concordant with demuxlet predictions and is highly consistent with the known truth in cell-hashing dataset.
Highlights
-
scSplit is an accurate, fast, and computationally efficient method for demultiplexing of individual cells from pooled samples from scRNA-seq.
-
The new tool can be used without needing sample genotypes prior to mixing.
-
Mainly designed for droplet-based scRNA-seq but can also be used for scRNA-seq data generated from other types of protocols.
-
The full article can be found at https://doi.org/10.1186/s13059-019-1852-7.
Publication
Genotype-free demultiplexing of pooled single-cell RNA-seq
Jun Xu, Caitlin Falconer, Quan Nguyen, Joanna Crawford, Brett D. McKinnon, Sally Mortlock, Anne Senabouth, Stacey Andersen, Han Sheng Chiu, Longda Jiang, Nathan J. Palpant, Jian Yang, Michael D. Mueller, Alex W. Hewitt, Alice Pébay, Grant W. Montgomery, Joseph E. Powell & Lachlan J.M Coin
Genome Biology volume 20, Article number: 290 (2019)
GIH team
Jun Xu's Twitter
Collaborators