Benchmarking of long read sequencing technologies and genome assembly tools
In brief
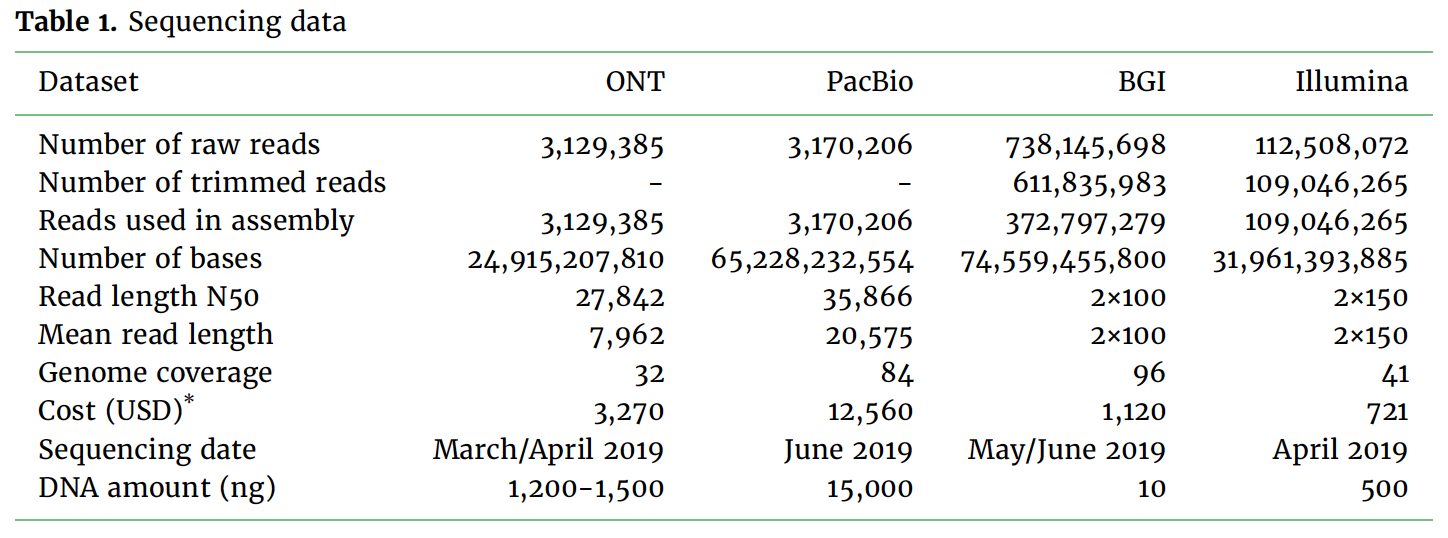
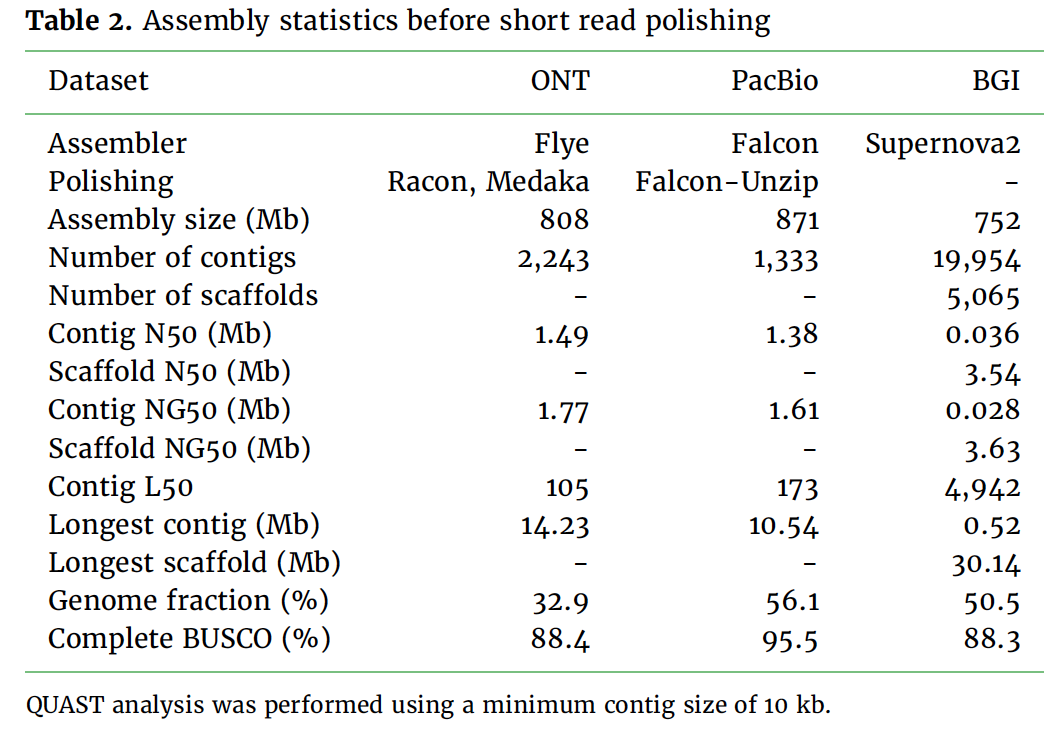
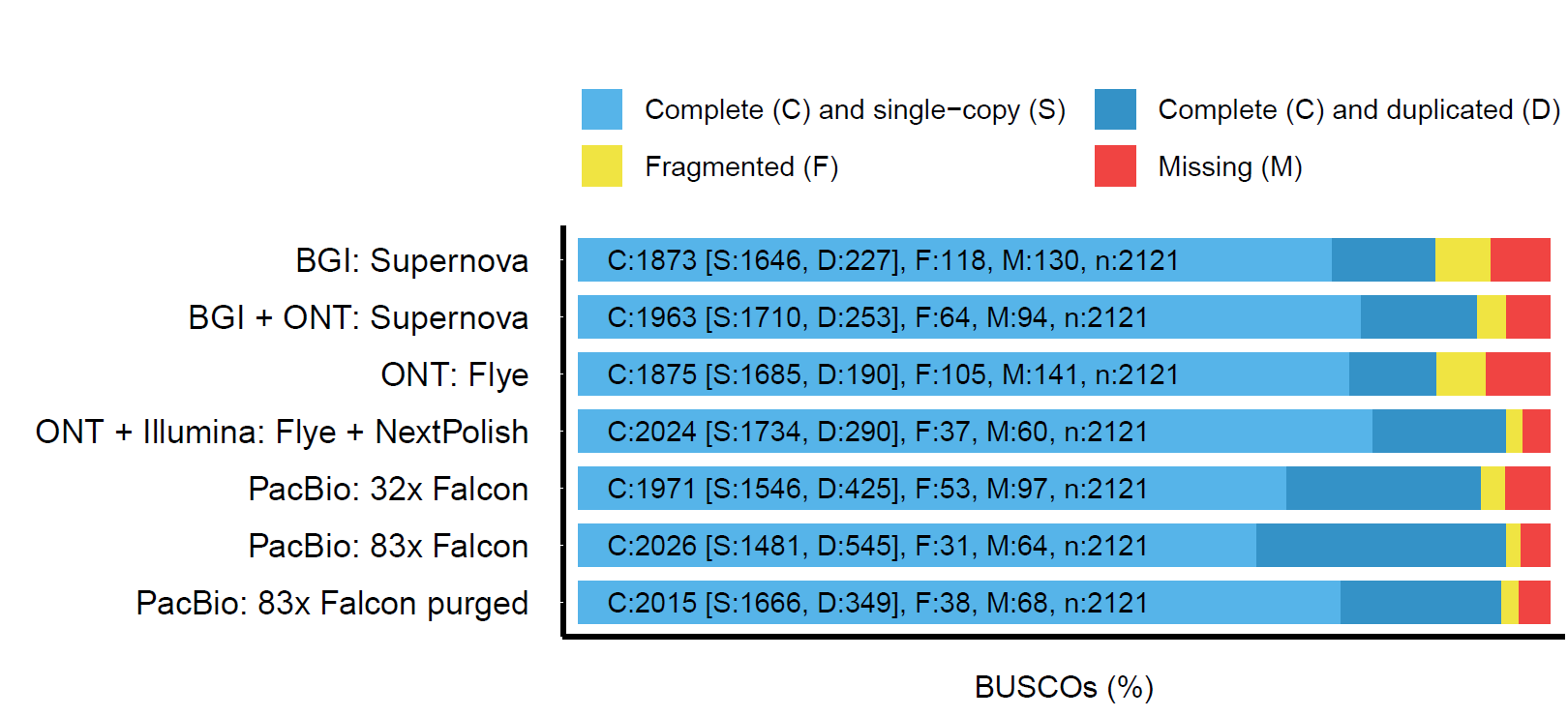
We report a comparison of three long read sequencing technologies applied to the de novo assembly of a plant genome, Macadamia jansenii. We have generated sequencing data using Pacific Biosciences (Sequel I), Oxford Nanopore Technologies (PromethION) and BGI (single-tube Long Fragment Read) technologies for the same sample. Several assemblers were benchmarked in the assembly of PacBio and Nanopore reads. The assemblies were compared for contiguity, accuracy and completeness as well as sequencing costs and DNA material requirements. Overall, the three long read technologies produced highly contiguous and complete genome assemblies of Macadamia jansenii. At the time of sequencing, the cost associated with each method was significantly different, but continuous improvements in technologies have resulted in greater accuracy, increased throughput and reduced costs.
Highlights
-
Three long read sequencing technologies were applied to the de novo assembly of a plant genome, Macadamia jansenii.
-
Several assemblers were benchmarked in the assembly of PacBio and Nanopore reads.
-
The assemblies were compared for contiguity, accuracy and completeness as well as sequencing costs and DNA material requirements.
-
A webinar presented by Valentine Murigneux and hosted by Australian Biocommons can be viewed at Webinar: Long read sequencing and assembly of Macadamia jansenii.
-
A webinar presented by Professor Lachlan Coin and hosted by BGI can be viewed at Webinar: Assembly of the Macadamia jansenii genome
Publications
Comparison of long-read methods for sequencing and assembly of a plant genome
Valentine Murigneux, Subash Kumar Rai, Agnelo Furtado, Timothy J C Bruxner, Wei Tian, Ivon Harliwong, Hanmin Wei, Bicheng Yang, Qianyu Ye, Ellis Anderson, Qing Mao, Radoje Drmanac, Ou Wang, Brock A Peters, Mengyang Xu, Pei Wu, Bruce Topp, Lachlan J M Coin, Robert J Henry
GigaScience, Volume 9, Issue 12, December 2020, giaa146, https://doi.org/10.1093/gigascience/giaa146
Improvements in the Sequencing and Assembly of Plant Genomes
Priyanka Sharma, Othman Aldossary, Bader Alsubaie, Ibrahim Al-Mssallem, Onkar Nath, Neena Mitter, Gabriel Margarido, Bruce Topp, Valentine Murigneux, Ardy Masouleh, Agnelo Furtado, Robert J Henry.
bioRxiv 2021.01.22.427724; doi: https://doi.org/10.1101/2021.01.22.427724
GIH Team
Collaborators